From basic biology to antibiotic targeting
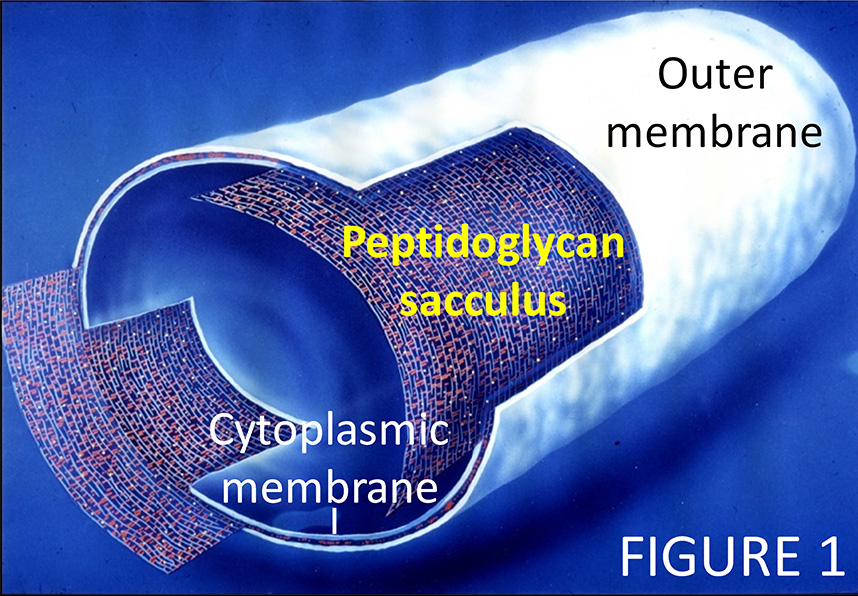 Bacteria build a robust cell envelope that enables them to inhabit all ecosystems on earth. Some disease-causing bacteria modify their cell wall to resist antibacterial components of our immune system. My group studies the bacterial cell envelope. For example, we want to gain understanding of how bacteria build and remodel their essential peptidoglycan sacculus (the "cell wall"), a net-like, stress-bearing layer in the cell envelope (1,2). In Gram-negative bacteria, the sacculus is sandwiched between the cytoplasmic membrane and an outer membrane, which protects the cell from many toxic molecules and antibacterial enzymes (Figure 1). Our research covers cell envelope topics in in model bacteria, for example Escherichia coli, and many other bacteria, including important pathogens, bacteria living in the environment and biotechnologically important bacteria.
Bacteria build a robust cell envelope that enables them to inhabit all ecosystems on earth. Some disease-causing bacteria modify their cell wall to resist antibacterial components of our immune system. My group studies the bacterial cell envelope. For example, we want to gain understanding of how bacteria build and remodel their essential peptidoglycan sacculus (the "cell wall"), a net-like, stress-bearing layer in the cell envelope (1,2). In Gram-negative bacteria, the sacculus is sandwiched between the cytoplasmic membrane and an outer membrane, which protects the cell from many toxic molecules and antibacterial enzymes (Figure 1). Our research covers cell envelope topics in in model bacteria, for example Escherichia coli, and many other bacteria, including important pathogens, bacteria living in the environment and biotechnologically important bacteria.
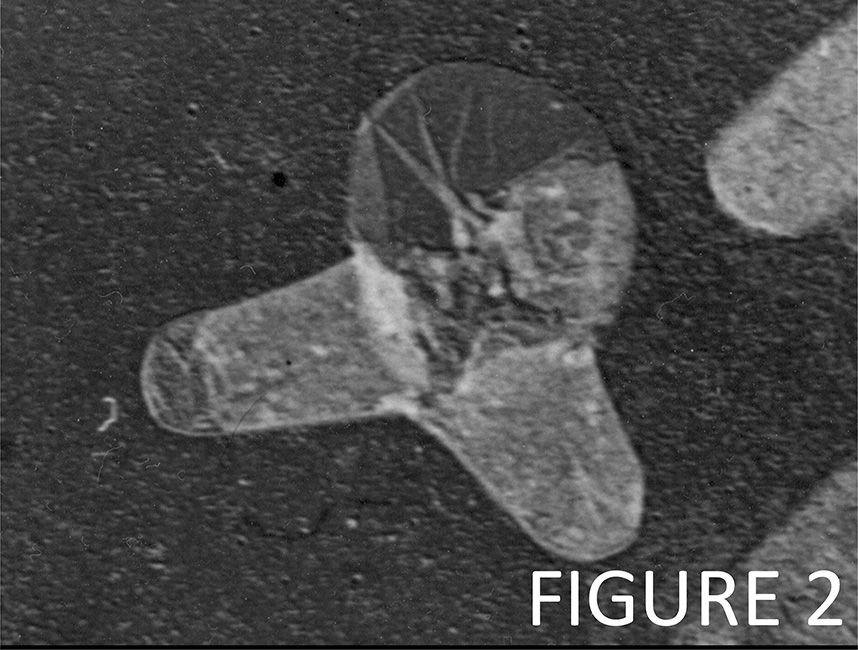 Our most successful antibiotics (e.g., beta-lactams like Pencillin) kill bacteria by blocking the synthesis of the peptidoglycan sacculus (PG). Figure 2 shows an E. coli cell that lysed by the action of Penicillin. We also develop assays to screen for novel antibiotics that target peptidoglycan synthesis and cell division, and we discover mechanisms of antimicrobial tolerance and resistance. We are also interested in how bacteria coordinate PG synthesis with the biogenesis of the other cell envelope layers and how bacteria maintain a robust cell envelope in different environments and under stress conditions.
Our most successful antibiotics (e.g., beta-lactams like Pencillin) kill bacteria by blocking the synthesis of the peptidoglycan sacculus (PG). Figure 2 shows an E. coli cell that lysed by the action of Penicillin. We also develop assays to screen for novel antibiotics that target peptidoglycan synthesis and cell division, and we discover mechanisms of antimicrobial tolerance and resistance. We are also interested in how bacteria coordinate PG synthesis with the biogenesis of the other cell envelope layers and how bacteria maintain a robust cell envelope in different environments and under stress conditions.
Group leader

Professor Waldemar Vollmer
Group Leader, Bacterial cell wall
+61 7 334 62055
w.vollmer@imb.uq.edu.au
UQ Experts Profile
Molecular mechanisms of PG synthesis
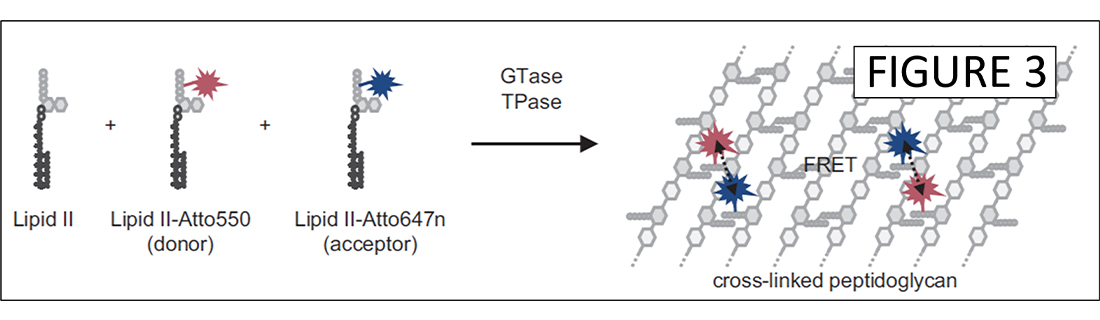 My laboratory has identified molecular interactions between different types of PG synthases (3,4). Synthases utilize lipid II precursor to polymerize glycan chains (GTase) and cross-link peptides (TPase), forming the net-like PG polymer. We have established in vitro PG synthesis assay, demonstrating the glycan chain polymerising and peptide cross-linking activities by key PG synthases (5,6). We recently developed a continuous PG synthesis assay for the analysis of membrane-reconstituted PG synthases (Figure 3) (7).
My laboratory has identified molecular interactions between different types of PG synthases (3,4). Synthases utilize lipid II precursor to polymerize glycan chains (GTase) and cross-link peptides (TPase), forming the net-like PG polymer. We have established in vitro PG synthesis assay, demonstrating the glycan chain polymerising and peptide cross-linking activities by key PG synthases (5,6). We recently developed a continuous PG synthesis assay for the analysis of membrane-reconstituted PG synthases (Figure 3) (7).
We have discovered with Nassos Typas and Carol Gross the lipoproteins LpoA and LpoB which activate major PG synthases (8). With Jean-Pierre Simorre we determined the structures of the activators and the interface between LpoB and PBP1B, providing the first model of a PG synthase - activator complex (9,10,11). We also discovered a mechanism by which the cell coordinates PG synthesis and outer membrane constriction in cell division (12) and showed how synthases are controlled by members of the divisome during pre-septal PG synthesis (13).
Robust cell envelope maintenance
E. coli has 18 known periplasmic autolysins (PG hydrolases) active during growth and cell division, and at least 6 PG carboxypeptidases for the regulation of PG synthesis. In 2016 we discovered a PG hydrolase that is required to maintain proper cell shape at acidic pH (14). With Tanneke den Blaauwen, Manuel Banzhaf and Nassos Typas we then discovered a network of interactions between different PG hydrolases and the adaptor protein NlpI (15) and with Petra Levin and Elizabeth Mueller deciphered the role of different PG synthases and hydrolases for growth under a range of environmental conditions (16,17). These findings support our model according to which the cell maintains sets of seemingly redundant PG enzymes to ensure that sufficient activity is available and regulated at a range of conditions the poorly buffered periplasm may encounter, allowing for robust cell envelope growth (18).
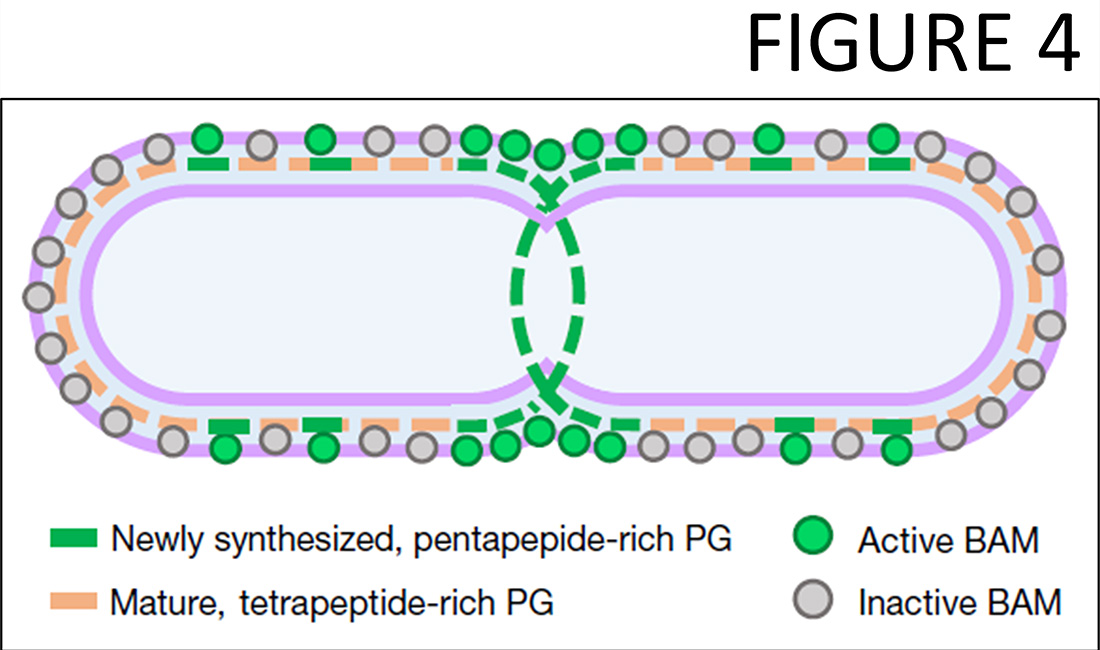 We recently began to study a novel research direction, the coordination between the biogenesis of the outer membrane with PG growth. Already our initial work with the group of Alessandra Polissi discovered a PG repair mechanism involving LD-transpeptidases that becomes essential when the biogenesis of the outer membrane is compromised (19) and showed that key LD-transpeptidases are inactivated by copper, preventing a PBP-bypass mechanisms for beta-lactam resistance (20). In 2022 we published with Colin Kleanthous and Gideon Mamou that the cell controls the essential outer membrane beta-barrel assembly machine (BAM) by the maturation state of the PG (21). Pentapeptide-rich PG, which occurs transiently at sites of PG growth, allows for BAM activity, whilst mature, tetrapeptide-rich PG inhibits BAM (Figure 4). This mechanism couples the BAM-mediated insertion of outer membrane (OM) proteins to PG growth sites, coordinating OM biogenesis with PG growth and maintaining a stable, functional OM around the PG layer.
We recently began to study a novel research direction, the coordination between the biogenesis of the outer membrane with PG growth. Already our initial work with the group of Alessandra Polissi discovered a PG repair mechanism involving LD-transpeptidases that becomes essential when the biogenesis of the outer membrane is compromised (19) and showed that key LD-transpeptidases are inactivated by copper, preventing a PBP-bypass mechanisms for beta-lactam resistance (20). In 2022 we published with Colin Kleanthous and Gideon Mamou that the cell controls the essential outer membrane beta-barrel assembly machine (BAM) by the maturation state of the PG (21). Pentapeptide-rich PG, which occurs transiently at sites of PG growth, allows for BAM activity, whilst mature, tetrapeptide-rich PG inhibits BAM (Figure 4). This mechanism couples the BAM-mediated insertion of outer membrane (OM) proteins to PG growth sites, coordinating OM biogenesis with PG growth and maintaining a stable, functional OM around the PG layer.
Bacterial cell shape and cell division
We are fortunate to have been collaborating with many other research groups on growth and cell shape of Gram-negative bacteria. For example, with Christine Jacobs-Wagner (Stanford) we have discovered FtsZ-dependent pre-septal PG synthesis (22) and a mechanism of mechanical control to generate cell curvature in Caulobacter crescentus (23), and established the mode of PG growth in Borrelia burgdorferi (24). With Nina Salama (Seattle) we have identified PG hydrolases required for helical cell shape and efficient stomach colonization in Helicobacter pylori (25). Subsequent work identified additional PG hydrolases participating in cell shape-generating pathways in H. pylori and, in collaboration with Erin Gaynor, Campylobacter jejuni (e.g., 26, 27). My group also contributed to the discovery of PG in chlamydia in collaboration with Grant Jensen and Martin Pilhofer (Caltech) (28). We have collaborated with the groups of Stephen Trent, Joe Boll and Mario Feldman on Acinetobacter baumannii and together we established the PG profile, discovered new PG modifications in the stationary phase, and showed the importance of LD-transpeptidases in colistin-resistant strains and PG recycling in carbapenem-resistance (e.g., 29, 30, 31).
The cell wall as target for antimicrobials and lysins
In the recent years, and based on our results from parts A-C, the focus of my group began to shift towards antimicrobial drug discovery. In collaboration with Cuong Voung (AiCuris Anti-infectives Cures GmbH, Wuppertal (Germany), and within a shared PhD studentship project, we identified lead inhibitors of the essential peptidoglycan synthase PBP3 (32). With Joen Luirink (Amsterdam) we identified novel inhibitors of the beta-barrel assembly machine (BAM) (33). We also work on antibacterial enzymes (lysins). For example, we collaborated with Joseph Mougous (Seattle) and Seemay Chou (San Francisco) on PG hydrolases that are secreted by type VI secretion system to target rival bacteria (34). Our subsequent work showed the wide distribution of these systems in bacteria (35) and provided a structural rationale for the high activity of these 'attacking hydrolases' (36). Some ticks and mites acquired PG hydrolase genes from bacterial origin during evolution (37) and use these hydrolases to control host skin bacteria during their blood meal (38). We have collaborated with Liz Sockett (Nottingham) and Andy Lovering (Birmingham) and identified PG hydrolases that Bdellovibrio uses to attack Gram-negative prey cells, showing these hydrolases are controlled by interaction with ankryn-like proteins and identified PG editing enzymes that generate the helical cell curvature (e.g., 39). With Jorge Galan (Yale) we discovered an LD-transpeptidase required for the secretion of typhoid toxin (40).
Novel cell wall modifiations, the cell wall architecture and new chemical tools
Within the last two decades and often within collaborations with others we discovered the founding genes encoding important cell wall enzymes in pathogenic bacteria including PG deacetylase (41), PG O-acetyltransferases (42), PG amidotransferase (43), several teichoic acid enzymes, e.g. the long-searched for enzyme that attaches anionic cell wall polymers (teichoic acids, capsular polysaccharides) to peptidoglycan (44) or phosphocholine esterase (45), and the first enzyme that is capable of detaching a protein from PG (46); homologues of all of these enzymes have been subsequently identified in many bacteria. Our expertise on the biochemistry of the cell wall structure has also helped our collaborators to characterise new chemical tools to study bacterial cell wall structure and biogenesis (e.g., 47, 48).
Our Approach
We discover molecular mechanisms in cell envelope biogenesis and use innovative biochemical assays to identify inhibitors of these. Our goal is to identify lead molecules for development of novel antibiotics that can cure infections caused by multi-resistant bacterial pathogens.
Research areas
Our research is focused on the bacterial cell wall and aims to answer the the following questions:
• How do bacteria enlarge their cell wall?
• How do they strengthen their cell envelope under stress conditions?
• How do antibiotics or components of our immune system attack the bacterial cell wall and how do some pathogenic bacteria resist these attacks?
Latest news
General enquiries
+61 7 3346 2222
imb@imb.uq.edu.au
Media enquiries
IMB fully supports UQ's Reconciliation Action Plan and is implementing actions within our institute.
Support us
Donate to research
100% of donations go to the cause


