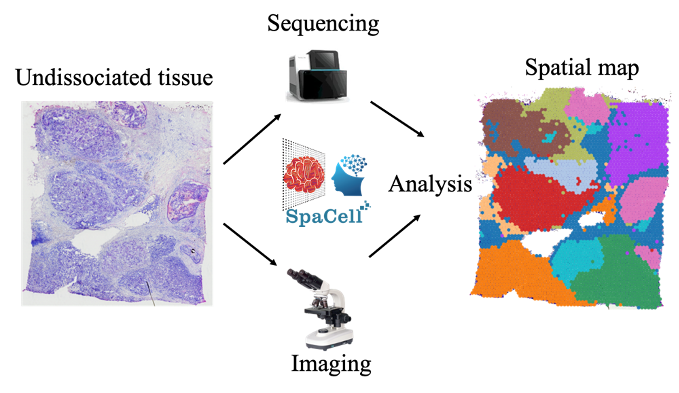
Spatial diagnosis of cancer tissue, including skin cancer, melanoma, and colorectal cancer

Two in three Australians have skin cancer before the age of 70. Each year, about 250,000 pigmented skin lesions are excised, which results in ~5% diagnosis of melanoma. Melanoma is the deadliest form of skin cancer and costs the health system over $272M annually. Early diagnosis of melanoma is challenging because malignant melanomas can mimic benign lesions that are overwhelmingly more common in the population.
Objective/mission (the vision)
We aim to test suspected cancer lesions in a fast, cost-effective, and accurate way. This is an essential, unmet need.
Research approach (the initiative)
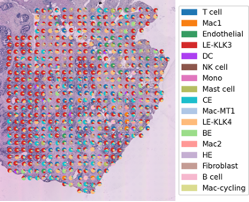
We will apply spatial transcriptomics as a revolutionary technology that can simultaneously capture tissue morphology and measure the expression of all genes without disassociating cells from their native tissue. The technology can, therefore, preserve the physiological spatial context. We will generate crucial understandings of in situ processes of cell-cell interactions, micro-metastasis, the level of immune infiltration, and the propensity of a lesion to metastasise. These are vital factors to detect and manage early melanoma. We will build computational models to identify locations within a tissue section that harbour clinically important cell types or suspected transformation/expansion/metastasis events.
Impacts and applications
We will build the next-generation analysis of pathological images for early melanoma detection, with potential applications to other cancer types. Our model results are to assist pathologists to assess images easier, faster and more accurately.
Partners/collaborators
In this project, we are collaborating with Prof Kiarash Khosroterani (UQDI), Dr Mitchell Stark (UQDI), Prof Peter Soyer (UQDI), Dr Jazmina and Prof Ian Frazer (UQDI), A/Prof Hanlee Ji (Stanford University), Dr HoJoon Lee (Stanford University)